
Computational Chemistry and Physics on the Nanoscale
CCPN 2009

Navigation
Construction, Visualization and Analysis of Structures in MD Simulations of Biomolecules
Augusto F. Oliveira
Computational simulations are becoming an important part of research on biomaterials and bionanotechnology. The increasing computer power, and the evolution of algorithms and computational methods have made possible to perform atomic level molecular dynamics simulations of systems containing several thousands of atoms in the nanosecond time scale.
However, as the simulated systems become larger and more heterogeneous, the construction of input coordinates and the visualization and analysis of the simulation results have also become more complex, requiring the processing of high amounts of data.
The objective of this course is to approach some of the software available for handling large molecular structures, such as proteins, simulated along large molecular dynamics trajectories. Special emphasis will be given to VMD, a very robust program visualization and analysis tool, empowered the Tcl scripting language.
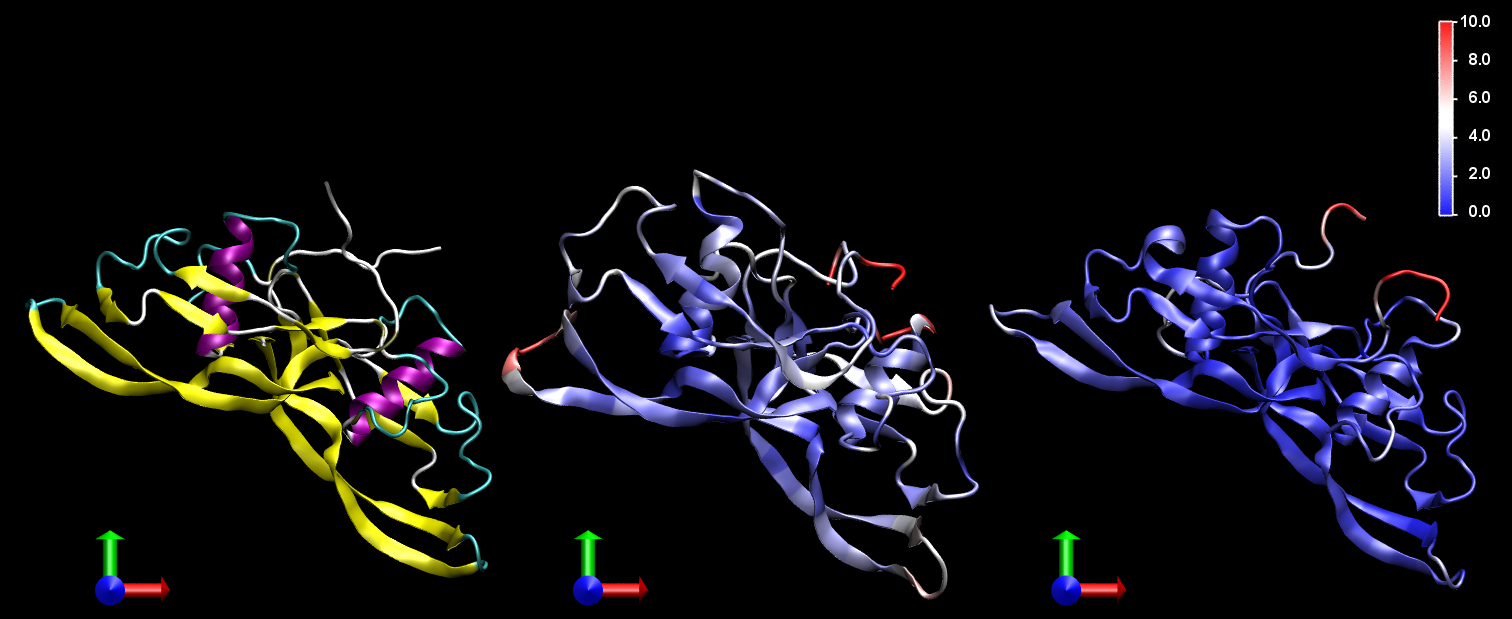
Structure of bone morphogenetic protein-2 (BMP-2). On left, the crystallographic structure, coloured according to its secondary structure. On center and right, the structures coloured by the root mean square deviation (RMSD) in Å, after 4 ns MD simulations in vacuum and water, respectively. The colour gradient helps to identify the flexible regions of the protein.
 General Information
General Information